Scientific workflows using R and git
Sara Mortara & Andrea Sánchez-Tapia
re.green | ¡liibre!
2022-07-07
today:
scientific workflows
good practices for writing scripts
- use our .Rproj structure to run a script
- generate outputs
commitof the day
scientific workflows
reproducibility - for you, collegues, and community
script based tools (
R,python)version control (
git)share methods, and protocols
peer review
our project structure
names and paths are essential to a reproducible workflow
project/ ├── .gitignore ├── data/ ├── docs/ ├── figs/ ├── R/ └── 02_importing_data.R ├── output/ ├── README.md └── .Rproj.Rproj defines the wd

.Rproj + git = <3

.Rproj + git = <3

.Rproj + git = <3
.Rproj + git = <3
workkflow: scripts
writing scripts
- Never create a single script with all the analysis
01_read_and_format_data.R02_diversity_analysis.R03_pca_analysis.R04_simulations.R...
writing scripts
- Never create a single script with all the analysis
01_read_and_format_data.R02_diversity_analysis.R03_pca_analysis.R04_simulations.R...
- Ideally, each script statrs reading a particular input/data and ends writing results
writing scripts
- Never create a single script with all the analysis
01_read_and_format_data.R02_diversity_analysis.R03_pca_analysis.R04_simulations.R...
Ideally, each script statrs reading a particular input/data and ends writing results
the next script can read raw data or results from previous scriptss.
example
R/01_data_clean.R
- reads
data/data_raw.csv - writes
data/data_processed.csv
example
R/01_data_clean.R
- reads
data/data_raw.csv - writes
data/data_processed.csv
R/02_diversity_analysis.R
- reads
data/data_processed.csv - writes
results/02_diversity.csvfigs/02_diversity.png
example
R/03_pca_analysis.R
- reads
data/data_processed.csv - writes
figs/03_pca.png
example
R/03_pca_analysis.R
- reads
data/data_processed.csv - writes
figs/03_pca.png
R/04_simulations.R
- reds
data/dados_processed.csv - saves
results/04_simulations.rdafigs/04_simulations.png
example
if an object is too large, or it takes too much time to process, it can be saved as an R object (.rda)
exemple:save(object, "./results/04_simulations.rda")
- following scripts can start loading these objects:
example: in the script 05_analysing_simulations.R
load("results/04_simulacoes.rda")
but never save the workspace!
organizing each script
Prints from swcarpentry.github.io/r-novice-inflammation/06-best-practices-R
each script
- a header containing who, how, when, where, and why METADATA

each script
- a header containing who, how, when, where, and why METADATA

- a part loading all needed packages from the begining with
library()*

each script
- reads needed data (empty workspace)

each script
- reads needed data (empty workspace)

- Coding a variable that will not change
each script
- reads needed data (empty workspace)

Coding a variable that will not change
Commenting every step
each script
- reads needed data (empty workspace)

Coding a variable that will not change
Commenting every step
Writing in the HD the result from each step
each script
the script must be able to be run in sequence from start to finish.
- No repetitions,
- No lines out of order
- No parentheses or non-closing calls (
png--->dev.off())
You should be able to erase the workspace mid-session and rebuild
Do not define functions inside the script. Put the functions in a separate script and folder
/fct/edit.Rand call viasource().
additional tips
use concise and informative names
a <-NO
do not use names already taken:
cor <-(color)cor()c <-If you copy and paste more than three times it's time to write a loop or a function
Getting back to the project
.├── 2022_scientific_computing_intro.Rproj├── data│ └── raw│ ├── cestes│ │ ├── comm.csv│ │ ├── coord.csv│ │ ├── envir.csv│ │ ├── README.md│ │ ├── splist.csv│ │ └── traits.csv│ └── portal_data_joined.csv├── docs│ └── scientific_workflows.Rmd├── figs├── output├── R│ ├── 01_intro.R│ └── 02_importing_data.R└── README.mdGetting back to git
CRLF vs LF
git config --global core.autocrlf false

More on this topic here
Getting back to git
git pull origin main
git add .gitignore
git add .
git commit -m "adding project's first strucure"
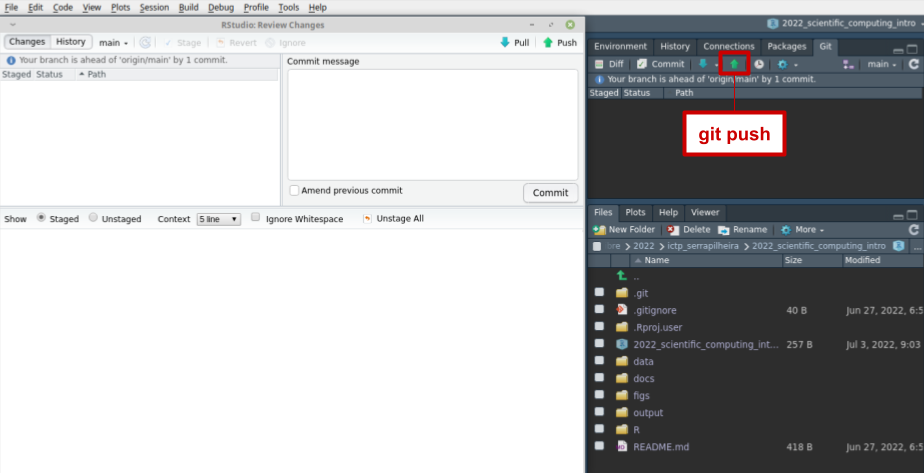
Running a script and generating outputs
git pull origin main
git add output/02_envir_summary.csv
git add figs/02_species_abundance.png
git commit -m "a very informative message about the scripts you're adding"
git push origin main
Creating a report
- Rmarkdown basic structure
run docs/scientific_workflow.Rmd
