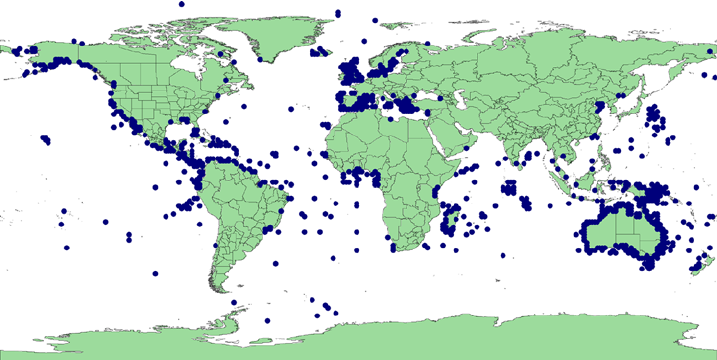
Biodiversity databases
Serrapilheira/ICTP-SAIFR Training Program in Quantitative Biology and Ecology
Andrea Sánchez-Tapia & Sara Mortara
27 July 2022
today
- Biodiversity databases: basic concepts
today
Biodiversity databases: basic concepts
DarwinCore standard
today
Biodiversity databases: basic concepts
DarwinCore standard
Biodiversity data cleaning
today
Biodiversity databases: basic concepts
DarwinCore standard
Biodiversity data cleaning
A workflow in R
1. Biodiversity databases: basic concepts
Biodiversity data
Museums, herbaria, collections


Who creates these data?
- Researchers: taxonomists, ecologists, field biologists
- Undergrad and grad school courses
- Amateur collectors, citizen science, using apps or not
Who creates these data?
- Researchers: taxonomists, ecologists, field biologists
- Undergrad and grad school courses
- Amateur collectors, citizen science, using apps or not
Who uses them?
- Researchers
- Curators
- The overall community (e.g., bird watchers)
Who creates these data?
- Researchers: taxonomists, ecologists, field biologists
- Undergrad and grad school courses
- Amateur collectors, citizen science, using apps or not
Who uses them?
- Researchers
- Curators
- The overall community (e.g., bird watchers)
- A massive amount of data
Who creates these data?
- Researchers: taxonomists, ecologists, field biologists
- Undergrad and grad school courses
- Amateur collectors, citizen science, using apps or not
Who uses them?
- Researchers
- Curators
- The overall community (e.g., bird watchers)
A massive amount of data
New research areas from the analysis and synthesis of these data: Biogeography, macroecology
Who creates these data?
- Researchers: taxonomists, ecologists, field biologists
- Undergrad and grad school courses
- Amateur collectors, citizen science, using apps or not
Who uses them?
- Researchers
- Curators
- The overall community (e.g., bird watchers)
A massive amount of data
New research areas from the analysis and synthesis of these data: Biogeography, macroecology
However: heterogeneous quality, inequality between countries and institutions
Biodiversity databases
GBIF (Global Biodiversity Information Facility)
Country-wise biodiversity information systems:
- 🇧🇷 speciesLink, Sistema de Informações sobre Biodiversidade Brasileira SiBBr
- 🇨🇴 Sistema de Información sobre Biodiversidad SibColombia
- 🇲🇽 Sistema Nacional de Información sobre Biodiversidad SNIB
- 🇦🇷 Sistema de Información de Biodiversidad SIB
Biodiversity databases
GBIF (Global Biodiversity Information Facility)
Country-wise biodiversity information systems:
- 🇧🇷 speciesLink, Sistema de Informações sobre Biodiversidade Brasileira SiBBr
- 🇨🇴 Sistema de Información sobre Biodiversidad SibColombia
- 🇲🇽 Sistema Nacional de Información sobre Biodiversidad SNIB
- 🇦🇷 Sistema de Información de Biodiversidad SIB
- Countries that participate via ministries, agencies
Biodiversity databases
GBIF (Global Biodiversity Information Facility)
Country-wise biodiversity information systems:
- 🇧🇷 speciesLink, Sistema de Informações sobre Biodiversidade Brasileira SiBBr
- 🇨🇴 Sistema de Información sobre Biodiversidad SibColombia
- 🇲🇽 Sistema Nacional de Información sobre Biodiversidad SNIB
- 🇦🇷 Sistema de Información de Biodiversidad SIB
Countries that participate via ministries, agencies
Other systems:
- Natureserve/ BISON in the USA
- iNaturalist
By taxonomic group
By taxonomic group
By taxonomic group
Transforming science through open data, software & reproducibility
Transforming science through open data, software & reproducibility
- R packages to make data available, data cleaning, APIs
Transforming science through open data, software & reproducibility
R packages to make data available, data cleaning, APIs
Package peer-review
Transforming science through open data, software & reproducibility
R packages to make data available, data cleaning, APIs
Package peer-review
Packages related to biodiversity data retrieval, manipulation, and standardization:
finch,rgbif,taxize,taxview...
Looking at GBIF data
Let's download the data of a tree species from South America Myrsine coriacea from the Primulaceae family.
library(rgbif)library(dplyr)species <- "Myrsine coriacea"occs <- occ_search(scientificName = species)names(occs)#glimpse(occs)#names(occs$data)Column names returned from gbif follow the DarwinCore standard (https://dwc.tdwg.org).
Possible fields
- Species (taxonomic information)
Possible fields
Species (taxonomic information)
Locality, coordinates, geographic
Possible fields
Species (taxonomic information)
Locality, coordinates, geographic
Collector notes and species attributes, IUCN status
Possible fields
Species (taxonomic information)
Locality, coordinates, geographic
Collector notes and species attributes, IUCN status
Collectors, determiners, and authors' names
2.DarwinCore Standard
DarwinCore
goal: facilitate the sharing of information about biological diversity by providing identifiers, labels, and definitions
maintained by a working group, adopted globally by GBIF and most collections, constant evolution
reference guide:
Taxon,Event,Identification,Location

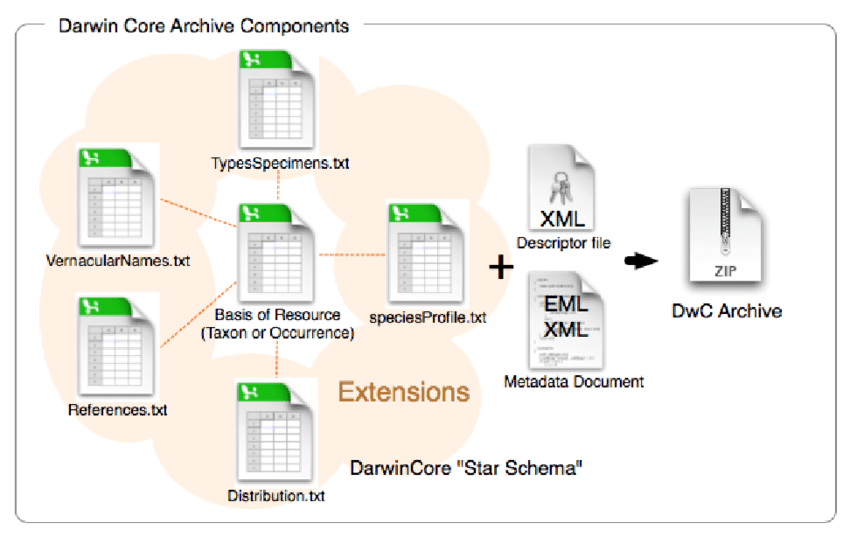
Multi-file DwC scheme
a ZIP file with:
- different data tables (relational databases)
- description file (XML)
- metadata files (EML Ecological Metadata Language)
Multi-file DwC scheme
a ZIP file with:
- different data tables (relational databases)
- description file (XML)
- metadata files (EML Ecological Metadata Language)
common columns for each table,
ID
- IPT is used to publish your data, either in one single table or extended: species lists, field work samplings etc. Datasets are given a permanent identifier and citation
Brazil Flora G (2020): Brazilian Flora 2020 project - Projeto Flora do Brasil 2020. v393.274. Instituto de Pesquisas Jardim Botanico do Rio de Janeiro. Dataset/Checklist. doi:10.15468/1mtkaw URL: http://ipt.jbrj.gov.br/jbrj/
49.343 species
136.314 taxa
Information about: distribution, habitat, endemism, references, synonyms
IPT in
R: package finch (Chamberlain 2020)
3. Data cleaning
Primary data are not clean or updated
Names and classifications change in time (synonyms):
International Commission on Zoological Nomenclature (ICZN), International Code of Botanical Nomenclature, International Association for Plant Taxonomy
The names of places: toponyms, we need gazetteers
Taxonomic identification errors, georreferencing, typographic/orthographic
Missing coordinates or localities
Imprecise coordinates, badly attributed
Taxonomic, nomenclatural erros
- Original vs. accepted name
Different taxonomic concepts / backbones, premisses, measure units.
New errors arising in processing and junction
Duplicate data vs. duplicatae

Data cleaning workflow
Check for
- format
- completeness
- sense (dimensions, ranges)
- outliers (geographic, temporary, environmental)
Good practices:
Always flag 🚩 additions, corrections, never modify original data
Document all steps -> reproducibility
Cite packages and data sources
Know your shortfalls
informed ignorance is a powerful research tool

References
Chapman, A. D. (2005). Principles and methods of data cleaning. GBIF.
Hortal, J., de Bello, F., Diniz-Filho, J. A. F., Lewinsohn, T. M., Lobo, J. M., & Ladle, R. J. (2015). Seven shortfalls that beset large-scale knowledge of biodiversity. Annual Review of Ecology, Evolution, and Systematics, 46, 523-549.
Yesson, C., Brewer, P. W., Sutton, T., Caithness, N., Pahwa, J. S., Burgess, M., ... & Culham, A. (2007). How global is the global biodiversity information facility?. PloS one, 2(11), e1124.
Wieczorek, J., Bloom, D., Guralnick, R., Blum, S., Döring, M., Giovanni, R., ... & Vieglais, D. (2012). Darwin Core: an evolving community-developed biodiversity data standard. PloS one, 7(1), e29715.